https://www.engr.colostate.edu/ets/keck-cluster/ under getting started
https://www.engr.colostate.edu/ets/get-connected/
https://www.engr.colostate.edu/ets/connect-to-the-keck-cluster/
tldr:
Mac/Linux
Open a terminal and type:
ssh <your user name>@ens-hpc.engr.colostate.edu , it will prompt for your ENS password.
Windows
- install PuTTy - ssh client used to connect
- install WinSCP - File transfer protocol client
- Open PuTTY
- Type
ens-hpc.engr.colostate.eduin the Hostname (IP address box), make sure the connection type is SSH. - Hit Open
- Enter ENS username and password
- Type
See the above ENS link to do graphical forwarding if you want.
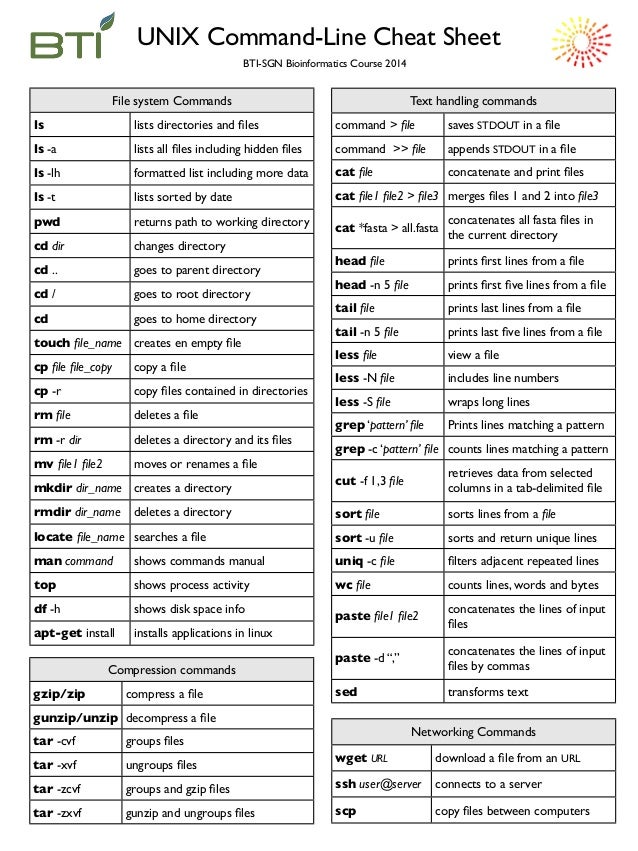
cd <directory> #change directory to requested location
. - current directory
.. - one directory up
ex: cd .. will move you one directory back up, cd ./folder/ will move you from your current directory into a folder
ls # prints all files within the current folder
mkdir <name> #makes a new directory in the current directory
Other useful ones to know:
TEXT EDITING Use vim or nano, these take a lot of practice to use effectively! Personally I prefer to write and edit my files locally then SCP them to the cluster to overwrite whats there.
For windows, you have a handy client WinSCP to do this for you, simply log in like you did with PuTTY and drag and drop files into desired locations.
For mac / linux: use the following commands in terminal
#Retreiving a file from the cluster: to a local directory
scp [email protected]:dir/file.txt /local/directory/
#sending a file to the cluster:
scp local/dir/file.txt username@yourIPaddress:/remote/directory/
#retreiving a whole directory from the cluster:
scp -r [email protected]:/remote/directory/ /local/directory/
#sending a whole directory to the cluster:
$ scp -r /local/directory/ username@yourIPaddress:/remote/directory/
This sets up a custom conda enviroment so you dont mess with the base install for your user using python enviroments
module load apps/anaconda3
conda init bash
Close session and log back in, command line will now have (base) in front of your >
conda create --name <name_of_project_goes_here> python=3.6
Here's a good list of conda commands: https://know.continuum.io/rs/387-XNW-688/images/conda-cheatsheet.pdf
Mostly never make changes to your base environment, resetting it is a pain. Everytime you log in, you must do:
conda activate <name_of_desired_env>
to use an environment, your (base) will change to (name_env)
conda info -e
will list all your current environments.
Now with that done you can use this as you would any conda terminal, pip install or conda install packages to customize what you are doing.
pip install <package>
conda install <package>
ENS guide: https://www.engr.colostate.edu/ets/keck-detailed-job-guide/
Setup a new anaconda environment:
conda create -n <name> gcc_linux-64
conda activate <name>
Now we have a new environment with a custom link to a compiler, we also need the c++ compilers so we need:
conda install -c anaconda gxx_linux-64
From here the following commands will install and compile rsnapsim in the local environment (this will be updated for conda install but for now its pip):
conda install eigen
conda install pip
pip install rsnapsim-ssa-cpp
pip install rsnapsim
To run a batch job you usually need 3 files: - The function you are parallelizing - a shell script wrapper (.sh) - an outer loop to call that wrapper while changing inputs You also need a few places to put outputs and files:
- /Logs/ (log files go here)
- /Errs/ (error files go here)
- /Results/ (outputs go here)
There's a sample job provided here in Example_rsnapsim_run
- run_rss_solver.py - the function we are calling to parallelize
- bash_call_rss_solver.sh - outer shell wrapper
- run_bactin_diff_kis.py - a loop to call "run_rss_solver.py" with 3 different initiation rates
You will have to change the python within bash_call_rss_solver.sh to your local python:
~/.conda/envs/<your environment name>/bin/python (There's a way to get around this I don't know it however, will add)
The batch job is submitted by python run_bactin_diff_kis.py which then will proceed to call bash_call_rss_solver.sh three times with three different initial rates, which in turn will call run_rss_solver.py on 3 threads on 1-3 nodes depending on availability.
seeing current jobs:
qstat # <--just your jobs
qstat -f # full cluster usage at the moment
qstat -u "*" # see all current users taking up cores
qstat will say your job is in one of a few different states:
qw - queue waiting, your job is waiting to be submitted once a node is free
t - transferring to a node
r - running
E / E - Some sort of error occured
s - suspended, something has gone wrong and needs to be trouble shooted
h - on hold something is wrong
typically anything on hold will need to be deleted with
qdel <job id> (job id is the first long digit long number when you hit qstat)
A special case: Eqw - Error in sending the job to a node, I usually get this when your command inside the shell script is wrong, i.e. the node you sent this too does not have access to python or matlab.
Eqw is annoying because it may not give a clear error message when you use qjob -j <jobid> | grep error to pull its error message.
for example of it getting the wrong matlab:
usr\local\bin\matlab -nosplash -nodisplay -r "matlabfun(); exit;"
This may not work but running module load apps/matlab-2020a then having this shell script will:
matlab -nosplash -nodisplay -r "matlabfun(); exit;"
the full qsub man page: https://www.univa.com/resources/files/univa_user_guide_univa__grid_engine_854.pdf
From within a conda enviroment on the HPC:
conda install jupyterlab #(if you need to install jupyter)
Then to log into an interactive session:
qlogin
conda activate <env_with_jupyter>
jupyter notebook --no-browser --ip=127.0.0.1 --port=<port number>
It should return you a link to copy paste Then locally you will have to open up an SSH tunnel by opening up a terminal or command prompt and typing:
ssh -N -L localhost:<port number>:localhost:<port number> [email protected]
It will prompt for your password, then from there copy paste the link the HPC gave you and you should have a local notebook running in your browser that is using the cluster. <port number> can be any 4 digit number, like 3333.
git clone https://github.com/Will-Raymond/keck_cluster_tutorial.git
Say we want to train a bunch of ffNN in parallel, we can do that by writing a python script that calls the GenericML class, the function we are going to call is train_ML_model.py it takes 3 inputs:
- type of model ('ffnn' or 'cnn' or 'knn' or 'rf')
- model name (str)
- model layer size if applicable, which in this case we are going to train ffNNs (list of layers)
- model save directory (str)
Remember the structure is the following:
[qsub Python loop] ---> [Bash call] ---> [Python/Matlab/commandline function]
fill in the following python script, run_ml_cluster.py, to call the qsub loop:
import subprocess
import time
<var1> = 'ffnn'
<var2> = 'example_ffnn'
<var4> = './ML_models/'
import os #make a directory if its not there
if not os.path.exists(<var4>):
os.makedirs(<var4>)
layers = [ [100,50,20,3], \ #the FFNN architectures to try!
[100,200,100,25,3], \
[40,200,300,100,3], \
[500,300,200,100,50,3]]
for <var3> in layers:
cmd = ' '.join( ['qsub','-q munsky.q@node* -cwd -o /dev/null -e Errs/',
'-v','','<var1>=%s' % (<var1>),
'-v','','<var2>=%s' % (<var2>),
'-v','','<var3>=%s' % (str(<var3>)),
'-v','','<var4>=%s' % (<var4>),
'bash_call_rss_solver.sh'] )
print(cmd)
subprocess.call( cmd, shell=True )
time.sleep(0.1) # delay for 0.1 seconds.
fill in the following shell script, bash_call_train_ml.sh:
~/.conda/envs/cc_env/bin/python [Python script] $[var1] $[var2] $[var3] > Logs/Log_ML_training_$[var1]_$[var2]_$[var3].txt
So our structure should be
run_ml_cluster.py ---> bash_call_train_ml.sh ---on nodes---> train_ML_model.py
we can now run this as
python run_ml_cluster.py