From our first two lessons, we are now equipped with enough fundamental programming skills to apply it to various steps in the data science workflow, which is a natural cycle that occurs in data analysis.
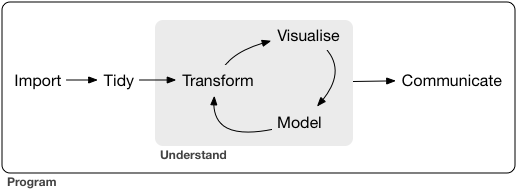 -
-For the rest of the course, we focus on Transform and Visualize with the assumption that our data is in a nice, “Tidy format”. First, we need to understand what it means for a data to be “Tidy”.
3.1 Tidy Data
 -A tidy dataframe. Image source: R for Data Science.
+A tidy dataframe. Image source: R for Data Science.
-A tidy dataframe. Image source: R for Data Science.
+A tidy dataframe. Image source: R for Data Science.
3.3.0.1 Let’s convert our impli
3.4 Summary Statistics
Now that your Dataframe has be transformed based on your scientific question, you can start doing some analysis on it! A common data science task is to examine summary statistics of a dataset, which summarizes the all the values from a variable in a numeric summary, such as mean, median, or mode.
-If we look at the data structre of a Dataframe’s column, it is called a Series. It has methods can compute summary statistics for us. Let’s take a look at a few popular examples:
+If we look at the data structure of a Dataframe’s column, it is actually not a List, but an object called Series. It has methods can compute summary statistics for us. Let’s take a look at a few popular examples:
-
@@ -418,25 +418,25 @@ 3.4 Summary Statistics
metadata.Age.mean()metadata.Age as a numeric valuemetadata.Age as a numeric SeriesComputes the mean value of the Age column.
Float (NumPy)
metadata['Age'].median()metadata['Age'] as a numeric valuemetadata['Age'] as a numeric SeriesComputes the median value of the Age column.
Float (NumPy)
metadata.Age.max()metadata.Age as a numeric valuemetadata.Age as a numeric SeriesComputes the max value of the Age column.
Float (NumPy)
metadata.OncotreeSubtype.value_counts()metadata.OncotreeSubtype as a Stringmetadata.OncotreeSubtype as a string SeriesCreates a frequency table of all unique elements in OncotreeSubtype column.
Series
@@ -479,18 +479,17 @@ 3.4 Summary Statistics
3.5 Simple data visualization
-We will dedicate extensive time later this course to talk about data visualization, but the Dataframe’s column, Series, has a method called .plot() that can help us make plots. The .plot() method will default make a line plot, but it is not necessary the plot style we want, so we can give the optional argument kind a String value to specify the plot style. We use it for making a histogram.
-The .plot() method also exists for Dataframes, in which you need to specify a plot using multiple columns. We use it for making a bar plot.
-
+We will dedicate extensive time later this course to talk about data visualization, but the Dataframe’s column, Series, has a method called .plot() that can help us make simple plots for one variable. The .plot() method will by default make a line plot, but it is not necessary the plot style we want, so we can give the optional argument kind a String value to specify the plot style. We use it for making a histogram or bar plot.
+
-
@@ -527,7 +526,36 @@ 3.5 Simple data visualizationmetadata.OncotreeLineage.value_counts().plot(kind = "bar")
plt.show()

-Notice here that we start with the column metadata.OncotreeLineage, and then we first use the method .value_counts() to get a Dataframe of a frequency table. Then, we take the frequency table Dataframe and use the .plot() method. It is quite common in Python to have multiple “chained” function calls, in which the output of .value_counts() is used for the input of .plot(). It takes a bit of time to get used to this!
+(The plt.figure() and plt.show() functions are used to render the plots on the website, but you don’t need to use it for your exercises - yet. We will discuss this in more detail during our week of data visualization.)
+
+3.5.0.1 Chained function calls
+Let’s look at our bar plot syntax more carefully. We start with the column metadata.OncotreeLineage, and then we first use the method .value_counts() to get a Series of a frequency table. Then, we take the frequency table Series and use the .plot() method.
+It is quite common in Python to have multiple “chained” function calls, in which the output of .value_counts() is used for the input of .plot() all in one line of code. It takes a bit of time to get used to this!
+Here’s another example of a chained function call, which looks quite complex, but let’s break it down:
+plt.figure()
+
+metadata.loc[metadata.AgeCategory == "Adult", ].OncotreeLineage.value_counts().plot(kind="bar")
+
+plt.show()
+
+
+- We first take the entire
metadata and do some subsetting, which outputs a Dataframe.
+- We access the
OncotreeLineage column, which outputs a Series.
+- We use the method
.value_counts(), which outputs a Series.
+- We make a plot out of it!
+
+We could have, alternatively, done this in several lines of code:
+plt.figure()
+
+metadata_subset = metadata.loc[metadata.AgeCategory == "Adult", ]
+metadata_subset_lineage = metadata_subset.OncotreeLineage
+lineage_freq = metadata_subset_lineage.value_counts()
+lineage_freq.plot(kind = "bar")
+
+plt.show()
+
+These are two different styles of code, but they do the exact same thing. It’s up to you to decide what is easier for you to understand.
+
3.6 Exercises
diff --git a/docs/no_toc/reference-keys.txt b/docs/no_toc/reference-keys.txt
index 231327f..e710f25 100644
--- a/docs/no_toc/reference-keys.txt
+++ b/docs/no_toc/reference-keys.txt
@@ -33,5 +33,6 @@ transform-what-do-you-want-to-do-with-this-dataframe
lets-convert-our-implicit-subsetting-criteria-into-code
summary-statistics
simple-data-visualization
+chained-function-calls
exercises-2
references
diff --git a/docs/no_toc/resources/images/03-data-wrangling1_files/figure-html/unnamed-chunk-13-5.png b/docs/no_toc/resources/images/03-data-wrangling1_files/figure-html/unnamed-chunk-13-5.png
new file mode 100644
index 0000000..9ef4a07
Binary files /dev/null and b/docs/no_toc/resources/images/03-data-wrangling1_files/figure-html/unnamed-chunk-13-5.png differ
diff --git a/docs/no_toc/resources/images/03-data-wrangling1_files/figure-html/unnamed-chunk-14-7.png b/docs/no_toc/resources/images/03-data-wrangling1_files/figure-html/unnamed-chunk-14-7.png
new file mode 100644
index 0000000..9ef4a07
Binary files /dev/null and b/docs/no_toc/resources/images/03-data-wrangling1_files/figure-html/unnamed-chunk-14-7.png differ
diff --git a/docs/no_toc/search_index.json b/docs/no_toc/search_index.json
index 4573e91..299b3db 100644
--- a/docs/no_toc/search_index.json
+++ b/docs/no_toc/search_index.json
@@ -1 +1 @@
-[["index.html", "Introduction to Python About this Course 0.1 Curriculum 0.2 Target Audience 0.3 Learning Objectives 0.4 Offerings", " Introduction to Python August, 2024 About this Course 0.1 Curriculum The course covers fundamentals of Python, a high-level programming language, and use it to wrangle data for analysis and visualization. 0.2 Target Audience The course is intended for researchers who want to learn coding for the first time with a data science application via the Python language. This course is also appropriate for folks who have explored data science or programming on their own and want to focus on some fundamentals. 0.3 Learning Objectives Analyze Tidy datasets in the Python programming language via data subsetting, joining, and transformations. Evaluate summary statistics and data visualization to understand scientific questions. Describe how the Python programming environment interpret complex expressions made out of functions, operations, and data structures, in a step-by-step way. Apply problem solving strategies to debug broken code. 0.4 Offerings This course is taught on a regular basis at Fred Hutch Cancer Center through the Data Science Lab. Announcements of course offering can be found here. "],["intro-to-computing.html", "Chapter 1 Intro to Computing 1.1 Goals of the course 1.2 What is a computer program? 1.3 A programming language has following elements: 1.4 Google Colab Setup 1.5 Grammar Structure 1: Evaluation of Expressions 1.6 Grammar Structure 2: Storing data types in the Variable Environment 1.7 Grammar Structure 3: Evaluation of Functions 1.8 Tips on writing your first code 1.9 Exercises", " Chapter 1 Intro to Computing Welcome to Introduction to Python! Each week, we cover a chapter, which consists of a lesson and exercise. In our first week together, we will look at big conceptual themes in programming, see how code is run, and learn some basic grammar structures of programming. 1.1 Goals of the course In the next 6 weeks, we will explore: Fundamental concepts in high-level programming languages (Python, R, Julia, etc.) that is transferable: How do programs run, and how do we solve problems using functions and data structures? Beginning of data science fundamentals: How do you translate your scientific question to a data wrangling problem and answer it? Data science workflow. Image source: R for Data Science. Find a nice balance between the two throughout the course: we will try to reproduce a figure from a scientific publication using new data. 1.2 What is a computer program? A sequence of instructions to manipulate data for the computer to execute. A series of translations: English <-> Programming Code for Interpreter <-> Machine Code for Central Processing Unit (CPU) We will focus on English <-> Programming Code for Python Interpreter in this class. More importantly: How we organize ideas <-> Instructing a computer to do something. 1.3 A programming language has following elements: Grammar structure to construct expressions; combining expressions to create more complex expressions Encapsulate complex expressions via functions to create modular and reusable tasks Encapsulate complex data via data structures to allow efficient manipulation of data 1.4 Google Colab Setup Google Colab is a Integrated Development Environment (IDE) on a web browser. Think about it as Microsoft Word to a plain text editor. It provides extra bells and whistles to using Python that is easier for the user. Let’s open up the KRAS analysis in Google Colab. If you are taking this course while it is in session, the project name is probably named “KRAS Demo” in your Google Classroom workspace. If you are taking this course on your own time, you can view it here. Today, we will pay close attention to: Python Console (“Executions”): Open it via View -> Executed code history. You give it one line of Python code, and the console executes that single line of code; you give it a single piece of instruction, and it executes it for you. Notebook: in the central panel of the website, you will see Python code interspersed with word document text. This is called a Python Notebook (other similar services include Jupyter Notebook, iPython Notebook), which has chunks of plain text and Python code, and it helps us understand better the code we are writing. Variable Environment: Open it by clicking on the “{x}” button on the left-hand panel. Often, your code will store information in the Variable Environment, so that information can be reused. For instance, we often load in data and store it in the Variable Environment, and use it throughout rest of your Python code. The first thing we will do is see the different ways we can run Python code. You can do the following: Type something into the Python Console (Execution) and click the arrow button, such as 2+2. The Python Console will run it and give you an output. Look through the Python Notebook, and when you see a chunk of Python Code, click the arrow button. It will copy the Python code chunk to the Python Console and run all of it. You will likely see variables created in the Variables panel as you load in and manipulate data. Run every single Python code chunk via Runtime -> Run all. Remember that the order that you run your code matters in programming. Your final product would be the result of Option 3, in which you run every Python code chunk from start to finish. However, sometimes it is nice to try out smaller parts of your code via Options 1 or 2. But you will be at risk of running your code out of order! To create your own content in the notebook, click on a section you want to insert content, and then click on “+ Code” or “+ Text” to add Python code or text, respectively. Python Notebook is great for data science work, because: It encourages reproducible data analysis, when you run your analysis from start to finish. It encourages excellent documentation, as you can have code, output from code, and prose combined together. It is flexible to use other programming languages, such as R. Now, we will get to the basics of programming grammar. 1.5 Grammar Structure 1: Evaluation of Expressions Expressions are be built out of operations or functions. Functions and operations take in data types, do something with them, and return another data type. We can combine multiple expressions together to form more complex expressions: an expression can have other expressions nested inside it. For instance, consider the following expressions entered to the Python Console: 18 + 21 ## 39 max(18, 21) ## 21 max(18 + 21, 65) ## 65 18 + (21 + 65) ## 104 len("ATCG") ## 4 Here, our input data types to the operation are integer in lines 1-4 and our input data type to the function is string in line 5. We will go over common data types shortly. Operations are just functions in hiding. We could have written: from operator import add add(18, 21) ## 39 add(18, add(21, 65)) ## 104 Remember that the Python language is supposed to help us understand what we are writing in code easily, lending to readable code. Therefore, it is sometimes useful to come up with operations that is easier to read. (Most functions in Python are stored in a collection of functions called modules that needs to be loaded. The import statement gives us permission to access the functions in the module “operator”.) 1.5.1 Data types Here are some common data types we will be using in this course. Data type name Data type shorthand Examples Integer int 2, 4 Float float 3.5, -34.1009 String str “hello”, “234-234-8594” Boolean bool True, False 1.5.2 Function machine schema A nice way to summarize this first grammar structure is using the function machine schema, way back from algebra class: Function machine from algebra class. Here are some aspects of this schema to pay attention to: A programmer should not need to know how the function or operation is implemented in order to use it - this emphasizes abstraction and modular thinking, a foundation in any programming language. A function can have different kinds of inputs and outputs - it doesn’t need to be numbers. In the len() function, the input is a String, and the output is an Integer. We will see increasingly complex functions with all sorts of different inputs and outputs. 1.6 Grammar Structure 2: Storing data types in the Variable Environment To build up a computer program, we need to store our returned data type from our expression somewhere for downstream use. We can assign a variable to it as follows: x = 18 + 21 If you enter this in the Console, you will see that in the Variable Environment, the variable x has a value of 39. 1.6.1 Execution rule for variable assignment Evaluate the expression to the right of =. Bind variable to the left of = to the resulting value. The variable is stored in the Variable Environment. The Variable Environment is where all the variables are stored, and can be used for an expression anytime once it is defined. Only one unique variable name can be defined. The variable is stored in the working memory of your computer, Random Access Memory (RAM). This is temporary memory storage on the computer that can be accessed quickly. Typically a personal computer has 8, 16, 32 Gigabytes of RAM. Look, now x can be reused downstream: x - 2 ## 37 y = x * 2 It is quite common for programmers to have to look up the data type of a variable while they are coding. To learn about the data type of a variable, use the type() function on any variable in Python: type(y) ## <class 'int'> We should give useful variable names so that we know what to expect! If you are working with numerical sales data, consider num_sales instead of y. 1.7 Grammar Structure 3: Evaluation of Functions Let’s look at functions a little bit more formally: A function has a function name, arguments, and returns a data type. 1.7.1 Execution rule for functions: Evaluate the function by its arguments if there’s any, and if the arguments are functions or contains operations, evaluate those functions or operations first. The output of functions is called the returned value. Often, we will use multiple functions in a nested way, and it is important to understand how the Python console understand the order of operation. We can also use paranthesis to change the order of operation. Think about what the Python is going to do step-by–step in the lines of code below: max(len("hello"), 4) ## 5 (len("pumpkin") - 8) * 2 ## -2 If we don’t know how to use a function, such as pow(), we can ask for help: ?pow pow(base, exp, mod=None) Equivalent to base**exp with 2 arguments or base**exp % mod with 3 arguments Some types, such as ints, are able to use a more efficient algorithm when invoked using the three argument form. This shows the function takes in three input arguments: base, exp, and mod=None. When an argument has an assigned value of mod=None, that means the input argument already has a value, and you don’t need to specify anything, unless you want to. The following ways are equivalent ways of using the pow() function: pow(2, 3) ## 8 pow(base=2, exp=3) ## 8 pow(exp=3, base=2) ## 8 but this will give you something different: pow(3, 2) ## 9 And there is an operational equivalent: 2 ** 3 ## 8 We will mostly look at functions with input arguments and return types in this course, but not all functions need to have input arguments and output return. Here are some varieties of functions to stretch your horizons. Function call What it takes in What it does Returns pow(a, b) integer a, integer b Raises a to the bth power. Integer print(x) any data type x Prints out the value of x to the console. None datetime.now() Nothing Gets the current time. String 1.8 Tips on writing your first code Computer = powerful + stupid Computers are excellent at doing something specific over and over again, but is extremely rigid and lack flexibility. Here are some tips that is helpful for beginners: Write incrementally, test often Check your assumptions, especially using new functions, operations, and new data types. Live environments are great for testing, but not great for reproducibility. Ask for help! To get more familiar with the errors Python gives you, take a look at this summary of Python error messages. 1.9 Exercises Exercise for week 1 can be found here. "],["working-with-data-structures.html", "Chapter 2 Working with data structures 2.1 Lists 2.2 Objects in Python 2.3 Dataframes 2.4 Exercises", " Chapter 2 Working with data structures In our second lesson, we start to look at two data structures, Lists and Dataframes, that can handle a large amount of data for analysis. 2.1 Lists In the first exercise, you started to explore data structures, which store information about data types. You explored lists, which is an ordered collection of data types or data structures. Each element of a list contains a data type or another data structure. We can now store a vast amount of information in a list, and assign it to a single variable. Even more, we can use operations and functions on a list, modifying many elements within the list at once! This makes analyzing data much more scalable and less repetitive. We create a list via the bracket [ ] operation. staff = ["chris", "ted", "jeff"] chrNum = [2, 3, 1, 2, 2] mixedList = [False, False, False, "A", "B", 92] 2.1.1 Subsetting lists To access an element of a list, you can use the bracket notation [ ] to access the elements of the list. We simply access an element via the “index” number - the location of the data within the list. Here’s the tricky thing about the index number: it starts at 0! 1st element of chrNum: chrNum[0] 2nd element of chrNum: chrNum[1] … 5th element of chrNum: chrNum[4] With subsetting, you can modify elements of a list or use the element of a list as part of an expression. 2.1.2 Subsetting multiple elements of lists Suppose you want to access multiple elements of a list, such as accessing the first three elements of chrNum. You would use the slice operator :, which specifies: the index number to start the index number to stop, plus one. If you want to access the first three elements of chrNum: chrNum[0:3] ## [2, 3, 1] The first element’s index number is 0, the third element’s index number is 2, plus 1, which is 3. If you want to access the second and third elements of chrNum: chrNum[1:3] ## [3, 1] If you want to access everything but the first three elements of chrNum: chrNum[3:len(chrNum)] ## [2, 2] where len(chrNum) is the length of the list. When the start or stop index is not specified, it implies that you are subsetting starting the from the beginning of the list or subsetting to the end of the list, respectively: chrNum[:3] ## [2, 3, 1] chrNum[3:] ## [2, 2] More discussion of list slicing can be found here. 2.2 Objects in Python The list data structure has an organization and functionality that metaphorically represents a pen-and-paper list in our physical world. Like a physical object, we have examined: What does it contain (in terms of data)? What can it do (in terms of operations and functions)? And if it “makes sense” to us, then it is well-designed. The list data structure we have been working with is an example of an Object. The definition of an object allows us to ask the questions above: what does it contain, and what can it do? It is an organizational tool for a collection of data and functions that we can relate to, like a physical object. Formally, an object contains the following: Value that holds the essential data for the object. Attributes that store additional data for the object. Functions called Methods that can be used on the object. This organizing structure on an object applies to pretty much all Python data types and data structures. Let’s see how this applies to the list: Value: the contents of the list, such as [2, 3, 4]. Attributes that store additional values: Not relevant for lists. Methods that can be used on the object: chrNum.count(2) counts the number of instances 2 appears as an element of chrNum. Object methods are functions that does something with the object you are using it on. You should think about chrNum.count(2) as a function that takes in chrNum and 2 as inputs. If you want to use the count function on list mixedList, you would use mixedList.count(x). Here are some more examples of methods with lists: Function method What it takes in What it does Returns chrNum.count(x) list chrNum, data type x Counts the number of instances x appears as an element of chrNum. Integer chrNum.append(x) list chrNum, data type x Appends x to the end of the chrNum. None (but chrNum is modified!) chrNum.sort() list chrNum Sorts chrNum by ascending order. None (but chrNum is modified!) chrNum.reverse() list chrNum Reverses the order of chrNum. None (but chrNum is modified!) 2.3 Dataframes A Dataframe is a two-dimensional data structure that stores data like a spreadsheet does. The Dataframe data structure is found within a Python module called “Pandas”. A Python module is an organized collection of functions and data structures. The import statement below gives us permission to access the “Pandas” module via the variable pd. To load in a Dataframe from existing spreadsheet data, we use the function pd.read_csv(): import pandas as pd metadata = pd.read_csv("classroom_data/metadata.csv") type(metadata) ## <class 'pandas.core.frame.DataFrame'> There is a similar function pd.read_excel() for loading in Excel spreadsheets. Let’s investigate the Dataframe as an object: What does a Dataframe contain (in terms of data)? What can a Dataframe do (in terms of operations and functions)? 2.3.1 What does a Dataframe contain (in terms of data)? We first take a look at the contents: metadata ## ModelID ... OncotreeLineage ## 0 ACH-000001 ... Ovary/Fallopian Tube ## 1 ACH-000002 ... Myeloid ## 2 ACH-000003 ... Bowel ## 3 ACH-000004 ... Myeloid ## 4 ACH-000005 ... Myeloid ## ... ... ... ... ## 1859 ACH-002968 ... Esophagus/Stomach ## 1860 ACH-002972 ... Esophagus/Stomach ## 1861 ACH-002979 ... Esophagus/Stomach ## 1862 ACH-002981 ... Esophagus/Stomach ## 1863 ACH-003071 ... Lung ## ## [1864 rows x 30 columns] It looks like there are 1864 rows and 30 columns in this Dataframe, and when we display it it shows some of the data. We can look at specific columns by looking at attributes via the dot operation. We can also look at the columns via the bracket operation. metadata.ModelID ## 0 ACH-000001 ## 1 ACH-000002 ## 2 ACH-000003 ## 3 ACH-000004 ## 4 ACH-000005 ## ... ## 1859 ACH-002968 ## 1860 ACH-002972 ## 1861 ACH-002979 ## 1862 ACH-002981 ## 1863 ACH-003071 ## Name: ModelID, Length: 1864, dtype: object metadata['ModelID'] ## 0 ACH-000001 ## 1 ACH-000002 ## 2 ACH-000003 ## 3 ACH-000004 ## 4 ACH-000005 ## ... ## 1859 ACH-002968 ## 1860 ACH-002972 ## 1861 ACH-002979 ## 1862 ACH-002981 ## 1863 ACH-003071 ## Name: ModelID, Length: 1864, dtype: object The names of all columns is stored as an attribute, which can be accessed via the dot operation. metadata.columns ## Index(['ModelID', 'PatientID', 'CellLineName', 'StrippedCellLineName', 'Age', ## 'SourceType', 'SangerModelID', 'RRID', 'DepmapModelType', 'AgeCategory', ## 'GrowthPattern', 'LegacyMolecularSubtype', 'PrimaryOrMetastasis', ## 'SampleCollectionSite', 'Sex', 'SourceDetail', 'LegacySubSubtype', ## 'CatalogNumber', 'CCLEName', 'COSMICID', 'PublicComments', ## 'WTSIMasterCellID', 'EngineeredModel', 'TreatmentStatus', ## 'OnboardedMedia', 'PlateCoating', 'OncotreeCode', 'OncotreeSubtype', ## 'OncotreePrimaryDisease', 'OncotreeLineage'], ## dtype='object') The number of rows and columns are also stored as an attribute: metadata.shape ## (1864, 30) 2.3.2 What can a Dataframe do (in terms of operations and functions)? We can use the head() and tail() functions to look at the first few rows and last few rows of metadata, respectively: metadata.head() ## ModelID PatientID ... OncotreePrimaryDisease OncotreeLineage ## 0 ACH-000001 PT-gj46wT ... Ovarian Epithelial Tumor Ovary/Fallopian Tube ## 1 ACH-000002 PT-5qa3uk ... Acute Myeloid Leukemia Myeloid ## 2 ACH-000003 PT-puKIyc ... Colorectal Adenocarcinoma Bowel ## 3 ACH-000004 PT-q4K2cp ... Acute Myeloid Leukemia Myeloid ## 4 ACH-000005 PT-q4K2cp ... Acute Myeloid Leukemia Myeloid ## ## [5 rows x 30 columns] metadata.tail() ## ModelID PatientID ... OncotreePrimaryDisease OncotreeLineage ## 1859 ACH-002968 PT-pjhrsc ... Esophagogastric Adenocarcinoma Esophagus/Stomach ## 1860 ACH-002972 PT-dkXZB1 ... Esophagogastric Adenocarcinoma Esophagus/Stomach ## 1861 ACH-002979 PT-lyHTzo ... Esophagogastric Adenocarcinoma Esophagus/Stomach ## 1862 ACH-002981 PT-Z9akXf ... Esophagogastric Adenocarcinoma Esophagus/Stomach ## 1863 ACH-003071 PT-LAGmLq ... Lung Neuroendocrine Tumor Lung ## ## [5 rows x 30 columns] Both of these functions (without input arguments) are considered as methods: they are functions that does something with the Dataframe you are using it on. You should think about metadata.head() as a function that takes in metadata as an input. If we had another Dataframe called my_data and you want to use the same function, you will have to say my_data.head(). 2.3.2.1 Subsetting Dataframes Perhaps the most important operation you will can do with Dataframes is subsetting them. There are two ways to do it. The first way is to subset by numerical indicies, exactly like how we did for lists. You will use the iloc and bracket operations, and you give two slices: one for the row, and one for the column. Let’s start with a small dataframe to see how it works before returning to metadata: df = pd.DataFrame(data={'status': ["treated", "untreated", "untreated", "discharged", "treated"], 'age_case': [25, 43, 21, 65, 7], 'age_control': [49, 20, 32, 25, 32]}) df ## status age_case age_control ## 0 treated 25 49 ## 1 untreated 43 20 ## 2 untreated 21 32 ## 3 discharged 65 25 ## 4 treated 7 32 Here is how the dataframe looks like with the row and column index numbers: Subset the second to fourth rows, and the first two columns: Now, back to metadata dataframe. Subset the first 5 rows, and first two columns: metadata.iloc[:5, :2] ## ModelID PatientID ## 0 ACH-000001 PT-gj46wT ## 1 ACH-000002 PT-5qa3uk ## 2 ACH-000003 PT-puKIyc ## 3 ACH-000004 PT-q4K2cp ## 4 ACH-000005 PT-q4K2cp If we want a custom slice that is not sequential, we can use an integer list. Subset the last 5 rows, and the 1st and 10 and 21th column: metadata.iloc[5:, [1, 10, 21]] ## PatientID GrowthPattern WTSIMasterCellID ## 5 PT-ej13Dz Suspension 2167.0 ## 6 PT-NOXwpH Adherent 569.0 ## 7 PT-fp8PeY Adherent 1806.0 ## 8 PT-puKIyc Adherent 2104.0 ## 9 PT-AR7W9o Adherent NaN ## ... ... ... ... ## 1859 PT-pjhrsc Organoid NaN ## 1860 PT-dkXZB1 Organoid NaN ## 1861 PT-lyHTzo Organoid NaN ## 1862 PT-Z9akXf Organoid NaN ## 1863 PT-LAGmLq Suspension NaN ## ## [1859 rows x 3 columns] When we subset via numerical indicies, it’s called explicit subsetting. This is a great way to start thinking about subsetting your dataframes for analysis, but explicit subsetting can lead to some inconsistencies in the long run. For instance, suppose your collaborator added a new cell line to the metadata and changed the order of the column. Then your code to subset the last 5 rows and the columns will get you a different answer once the spreadsheet is changed. The second way is to subset by the column name and comparison operators, also known as implicit subsetting. This is much more robust in data analysis practice. You will learn about it next week! 2.4 Exercises Exercise for week 2 can be found here. "],["data-wrangling-part-1.html", "Chapter 3 Data Wrangling, Part 1 3.1 Tidy Data 3.2 Our working Tidy Data: DepMap Project 3.3 Transform: “What do you want to do with this Dataframe”? 3.4 Summary Statistics 3.5 Simple data visualization 3.6 Exercises", " Chapter 3 Data Wrangling, Part 1 From our first two lessons, we are now equipped with enough fundamental programming skills to apply it to various steps in the data science workflow, which is a natural cycle that occurs in data analysis. Data science workflow. Image source: R for Data Science. For the rest of the course, we focus on Transform and Visualize with the assumption that our data is in a nice, “Tidy format”. First, we need to understand what it means for a data to be “Tidy”. 3.1 Tidy Data Here, we describe a standard of organizing data. It is important to have standards, as it facilitates a consistent way of thinking about data organization and building tools (functions) that make use of that standard. The principles of tidy data, developed by Hadley Wickham: Each variable must have its own column. Each observation must have its own row. Each value must have its own cell. If you want to be technical about what variables and observations are, Hadley Wickham describes: A variable contains all values that measure the same underlying attribute (like height, temperature, duration) across units. An observation contains all values measured on the same unit (like a person, or a day, or a race) across attributes. A tidy dataframe. Image source: R for Data Science. 3.2 Our working Tidy Data: DepMap Project The Dependency Map project is a multi-omics profiling of cancer cell lines combined with functional assays such as CRISPR and drug sensitivity to help identify cancer vulnerabilities and drug targets. Here are some of the data that we have public access to. We have been looking at the metadata since last session. Metadata Somatic mutations Gene expression Drug sensitivity CRISPR knockout and more… Let’s load these datasets in, and see how these datasets fit the definition of Tidy data: import pandas as pd metadata = pd.read_csv("classroom_data/metadata.csv") mutation = pd.read_csv("classroom_data/mutation.csv") expression = pd.read_csv("classroom_data/expression.csv") metadata.head() ## ModelID PatientID ... OncotreePrimaryDisease OncotreeLineage ## 0 ACH-000001 PT-gj46wT ... Ovarian Epithelial Tumor Ovary/Fallopian Tube ## 1 ACH-000002 PT-5qa3uk ... Acute Myeloid Leukemia Myeloid ## 2 ACH-000003 PT-puKIyc ... Colorectal Adenocarcinoma Bowel ## 3 ACH-000004 PT-q4K2cp ... Acute Myeloid Leukemia Myeloid ## 4 ACH-000005 PT-q4K2cp ... Acute Myeloid Leukemia Myeloid ## ## [5 rows x 30 columns] mutation.head() ## ModelID CACNA1D_Mut CYP2D6_Mut ... CCDC28A_Mut C1orf194_Mut U2AF1_Mut ## 0 ACH-000001 False False ... False False False ## 1 ACH-000002 False False ... False False False ## 2 ACH-000004 False False ... False False False ## 3 ACH-000005 False False ... False False False ## 4 ACH-000006 False False ... False False False ## ## [5 rows x 540 columns] expression.head() ## ModelID ENPP4_Exp CREBBP_Exp ... OR5D13_Exp C2orf81_Exp OR8S1_Exp ## 0 ACH-001113 2.280956 4.094236 ... 0.0 1.726831 0.0 ## 1 ACH-001289 3.622930 3.606442 ... 0.0 0.790772 0.0 ## 2 ACH-001339 0.790772 2.970854 ... 0.0 0.575312 0.0 ## 3 ACH-001538 3.485427 2.801159 ... 0.0 1.077243 0.0 ## 4 ACH-000242 0.879706 3.327687 ... 0.0 0.722466 0.0 ## ## [5 rows x 536 columns] Dataframe The observation is Some variables are Some values are metadata Cell line ModelID, Age, OncotreeLineage “ACH-000001”, 60, “Myeloid” expression Cell line KRAS_Exp 2.4, .3 mutation Cell line KRAS_Mut TRUE, FALSE 3.3 Transform: “What do you want to do with this Dataframe”? Remember that a major theme of the course is about: How we organize ideas <-> Instructing a computer to do something. Until now, we haven’t focused too much on how we organize our scientific ideas to interact with what we can do with code. Let’s pivot to write our code driven by our scientific curiosity. After we are sure that we are working with Tidy data, we can ponder how we want to transform our data that satisfies our scientific question. We will look at several ways we can transform Tidy data, starting with subsetting columns and rows. Here’s a starting prompt: In the metadata dataframe, which rows would you subset for and columns would you subset for that relate to a scientific question? We have been using explicit subsetting with numerical indicies, such as “I want to filter for rows 20-50 and select columns 2 and 8”. We are now going to switch to implicit subsetting in which we describe the subsetting criteria via comparision operators and column names, such as: “I want to subset for rows such that the OncotreeLineage is breast cancer and subset for columns Age and Sex.” Notice that when we subset for rows in an implicit way, we formulate our criteria in terms of the columns.This is because we are guaranteed to have column names in Dataframes, but not row names. 3.3.0.1 Let’s convert our implicit subsetting criteria into code! To subset for rows implicitly, we will use the conditional operators on Dataframe columns you used in Exercise 2. To formulate a conditional operator expression that OncotreeLineage is breast cancer: metadata['OncotreeLineage'] == "Lung" ## 0 False ## 1 False ## 2 False ## 3 False ## 4 False ## ... ## 1859 False ## 1860 False ## 1861 False ## 1862 False ## 1863 True ## Name: OncotreeLineage, Length: 1864, dtype: bool Then, we will use the .loc operation (which is different than .iloc operation!) and subsetting brackets to subset rows and columns Age and Sex at the same time: metadata.loc[metadata['OncotreeLineage'] == "Lung", ["Age", "Sex"]] ## Age Sex ## 10 39.0 Female ## 13 44.0 Male ## 19 55.0 Female ## 27 39.0 Female ## 28 45.0 Male ## ... ... ... ## 1745 52.0 Male ## 1819 84.0 Male ## 1820 57.0 Female ## 1822 53.0 Male ## 1863 62.0 Male ## ## [241 rows x 2 columns] What’s going on here? The first component of the subset, metadata['OncotreeLineage'] == \"Lung\", subsets for the rows. It gives us a column of True and False values, and we keep rows that correspond to True values. Then, we specify the column names we want to subset for via a list. Here’s another example: df = pd.DataFrame(data={'status': ["treated", "untreated", "untreated", "discharged", "treated"], 'age_case': [25, 43, 21, 65, 7], 'age_control': [49, 20, 32, 25, 32]}) df ## status age_case age_control ## 0 treated 25 49 ## 1 untreated 43 20 ## 2 untreated 21 32 ## 3 discharged 65 25 ## 4 treated 7 32 “I want to subset for rows such that the status is”treated” and subset for columns status and age_case.” df.loc[df.status == "treated", ["status", "age_case"]] ## status age_case ## 0 treated 25 ## 4 treated 7 3.4 Summary Statistics Now that your Dataframe has be transformed based on your scientific question, you can start doing some analysis on it! A common data science task is to examine summary statistics of a dataset, which summarizes the all the values from a variable in a numeric summary, such as mean, median, or mode. If we look at the data structre of a Dataframe’s column, it is called a Series. It has methods can compute summary statistics for us. Let’s take a look at a few popular examples: Function method What it takes in What it does Returns metadata.Age.mean() metadata.Age as a numeric value Computes the mean value of the Age column. Float (NumPy) metadata['Age'].median() metadata['Age'] as a numeric value Computes the median value of the Age column. Float (NumPy) metadata.Age.max() metadata.Age as a numeric value Computes the max value of the Age column. Float (NumPy) metadata.OncotreeSubtype.value_counts() metadata.OncotreeSubtype as a String Creates a frequency table of all unique elements in OncotreeSubtype column. Series Let’s try it out, with some nice print formatting: print("Mean value of Age column:", metadata['Age'].mean()) ## Mean value of Age column: 47.45187165775401 print("Frequency of column", metadata.OncotreeLineage.value_counts()) ## Frequency of column OncotreeLineage ## Lung 241 ## Lymphoid 209 ## CNS/Brain 123 ## Skin 118 ## Esophagus/Stomach 95 ## Breast 92 ## Bowel 87 ## Head and Neck 81 ## Myeloid 77 ## Bone 75 ## Ovary/Fallopian Tube 74 ## Pancreas 65 ## Kidney 64 ## Peripheral Nervous System 55 ## Soft Tissue 54 ## Uterus 41 ## Fibroblast 41 ## Biliary Tract 40 ## Bladder/Urinary Tract 39 ## Normal 39 ## Pleura 35 ## Liver 28 ## Cervix 25 ## Eye 19 ## Thyroid 18 ## Prostate 14 ## Vulva/Vagina 5 ## Ampulla of Vater 4 ## Testis 4 ## Adrenal Gland 1 ## Other 1 ## Name: count, dtype: int64 (Notice that the output of some of these methods are Float (NumPy). This refers to a Python Object called NumPy that is extremely popular for scientific computing, but we’re not focused on that in this course.) 3.5 Simple data visualization We will dedicate extensive time later this course to talk about data visualization, but the Dataframe’s column, Series, has a method called .plot() that can help us make plots. The .plot() method will default make a line plot, but it is not necessary the plot style we want, so we can give the optional argument kind a String value to specify the plot style. We use it for making a histogram. The .plot() method also exists for Dataframes, in which you need to specify a plot using multiple columns. We use it for making a bar plot. Plot style Useful for kind = Code Histogram Numerics “hist” metadata.Age.plot(kind = \"hist\") Bar plot Strings “bar” metadata.OncotreeSubtype.value_counts().plot(kind = \"bar\") Let’s look at a histogram: import matplotlib.pyplot as plt plt.figure() metadata.Age.plot(kind = "hist") plt.show() Let’s look at a bar plot: plt.figure() metadata.OncotreeLineage.value_counts().plot(kind = "bar") plt.show() Notice here that we start with the column metadata.OncotreeLineage, and then we first use the method .value_counts() to get a Dataframe of a frequency table. Then, we take the frequency table Dataframe and use the .plot() method. It is quite common in Python to have multiple “chained” function calls, in which the output of .value_counts() is used for the input of .plot(). It takes a bit of time to get used to this! 3.6 Exercises Exercise for week 3 can be found here. "],["about-the-authors.html", "About the Authors", " About the Authors These credits are based on our course contributors table guidelines. Credits Names Pedagogy Lead Content Instructor(s) FirstName LastName Lecturer(s) (include chapter name/link in parentheses if only for specific chapters) - make new line if more than one chapter involved Delivered the course in some way - video or audio Content Author(s) (include chapter name/link in parentheses if only for specific chapters) - make new line if more than one chapter involved If any other authors besides lead instructor Content Contributor(s) (include section name/link in parentheses) - make new line if more than one section involved Wrote less than a chapter Content Editor(s)/Reviewer(s) Checked your content Content Director(s) Helped guide the content direction Content Consultants (include chapter name/link in parentheses or word “General”) - make new line if more than one chapter involved Gave high level advice on content Acknowledgments Gave small assistance to content but not to the level of consulting Production Content Publisher(s) Helped with publishing platform Content Publishing Reviewer(s) Reviewed overall content and aesthetics on publishing platform Technical Course Publishing Engineer(s) Helped with the code for the technical aspects related to the specific course generation Template Publishing Engineers Candace Savonen, Carrie Wright, Ava Hoffman Publishing Maintenance Engineer Candace Savonen Technical Publishing Stylists Carrie Wright, Ava Hoffman, Candace Savonen Package Developers (ottrpal) Candace Savonen, John Muschelli, Carrie Wright Art and Design Illustrator(s) Created graphics for the course Figure Artist(s) Created figures/plots for course Videographer(s) Filmed videos Videography Editor(s) Edited film Audiographer(s) Recorded audio Audiography Editor(s) Edited audio recordings Funding Funder(s) Institution/individual who funded course including grant number Funding Staff Staff members who help with funding ## ─ Session info ─────────────────────────────────────────────────────────────── ## setting value ## version R version 4.3.2 (2023-10-31) ## os Ubuntu 22.04.4 LTS ## system x86_64, linux-gnu ## ui X11 ## language (EN) ## collate en_US.UTF-8 ## ctype en_US.UTF-8 ## tz Etc/UTC ## date 2024-08-21 ## pandoc 3.1.1 @ /usr/local/bin/ (via rmarkdown) ## ## ─ Packages ─────────────────────────────────────────────────────────────────── ## package * version date (UTC) lib source ## askpass 1.2.0 2023-09-03 [1] RSPM (R 4.3.0) ## bookdown 0.39.1 2024-06-11 [1] Github (rstudio/bookdown@f244cf1) ## bslib 0.6.1 2023-11-28 [1] RSPM (R 4.3.0) ## cachem 1.0.8 2023-05-01 [1] RSPM (R 4.3.0) ## cli 3.6.2 2023-12-11 [1] RSPM (R 4.3.0) ## devtools 2.4.5 2022-10-11 [1] RSPM (R 4.3.0) ## digest 0.6.34 2024-01-11 [1] RSPM (R 4.3.0) ## ellipsis 0.3.2 2021-04-29 [1] RSPM (R 4.3.0) ## evaluate 0.23 2023-11-01 [1] RSPM (R 4.3.0) ## fansi 1.0.6 2023-12-08 [1] RSPM (R 4.3.0) ## fastmap 1.1.1 2023-02-24 [1] RSPM (R 4.3.0) ## fs 1.6.3 2023-07-20 [1] RSPM (R 4.3.0) ## glue 1.7.0 2024-01-09 [1] RSPM (R 4.3.0) ## hms 1.1.3 2023-03-21 [1] RSPM (R 4.3.0) ## htmltools 0.5.7 2023-11-03 [1] RSPM (R 4.3.0) ## htmlwidgets 1.6.4 2023-12-06 [1] RSPM (R 4.3.0) ## httpuv 1.6.14 2024-01-26 [1] RSPM (R 4.3.0) ## httr 1.4.7 2023-08-15 [1] RSPM (R 4.3.0) ## jquerylib 0.1.4 2021-04-26 [1] RSPM (R 4.3.0) ## jsonlite 1.8.8 2023-12-04 [1] RSPM (R 4.3.0) ## knitr 1.47.3 2024-06-11 [1] Github (yihui/knitr@e1edd34) ## later 1.3.2 2023-12-06 [1] RSPM (R 4.3.0) ## lifecycle 1.0.4 2023-11-07 [1] RSPM (R 4.3.0) ## magrittr 2.0.3 2022-03-30 [1] RSPM (R 4.3.0) ## memoise 2.0.1 2021-11-26 [1] RSPM (R 4.3.0) ## mime 0.12 2021-09-28 [1] RSPM (R 4.3.0) ## miniUI 0.1.1.1 2018-05-18 [1] RSPM (R 4.3.0) ## openssl 2.1.1 2023-09-25 [1] RSPM (R 4.3.0) ## ottrpal 1.2.1 2024-06-11 [1] Github (jhudsl/ottrpal@828539f) ## pillar 1.9.0 2023-03-22 [1] RSPM (R 4.3.0) ## pkgbuild 1.4.3 2023-12-10 [1] RSPM (R 4.3.0) ## pkgconfig 2.0.3 2019-09-22 [1] RSPM (R 4.3.0) ## pkgload 1.3.4 2024-01-16 [1] RSPM (R 4.3.0) ## profvis 0.3.8 2023-05-02 [1] RSPM (R 4.3.0) ## promises 1.2.1 2023-08-10 [1] RSPM (R 4.3.0) ## purrr 1.0.2 2023-08-10 [1] RSPM (R 4.3.0) ## R6 2.5.1 2021-08-19 [1] RSPM (R 4.3.0) ## Rcpp 1.0.12 2024-01-09 [1] RSPM (R 4.3.0) ## readr 2.1.5 2024-01-10 [1] RSPM (R 4.3.0) ## remotes 2.4.2.1 2023-07-18 [1] RSPM (R 4.3.0) ## rlang 1.1.4 2024-06-04 [1] CRAN (R 4.3.2) ## rmarkdown 2.27.1 2024-06-11 [1] Github (rstudio/rmarkdown@e1c93a9) ## sass 0.4.8 2023-12-06 [1] RSPM (R 4.3.0) ## sessioninfo 1.2.2 2021-12-06 [1] RSPM (R 4.3.0) ## shiny 1.8.0 2023-11-17 [1] RSPM (R 4.3.0) ## stringi 1.8.3 2023-12-11 [1] RSPM (R 4.3.0) ## stringr 1.5.1 2023-11-14 [1] RSPM (R 4.3.0) ## tibble 3.2.1 2023-03-20 [1] CRAN (R 4.3.2) ## tzdb 0.4.0 2023-05-12 [1] RSPM (R 4.3.0) ## urlchecker 1.0.1 2021-11-30 [1] RSPM (R 4.3.0) ## usethis 2.2.3 2024-02-19 [1] RSPM (R 4.3.0) ## utf8 1.2.4 2023-10-22 [1] RSPM (R 4.3.0) ## vctrs 0.6.5 2023-12-01 [1] RSPM (R 4.3.0) ## xfun 0.44.4 2024-06-11 [1] Github (yihui/xfun@9da62cc) ## xml2 1.3.6 2023-12-04 [1] RSPM (R 4.3.0) ## xtable 1.8-4 2019-04-21 [1] RSPM (R 4.3.0) ## yaml 2.3.8 2023-12-11 [1] RSPM (R 4.3.0) ## ## [1] /usr/local/lib/R/site-library ## [2] /usr/local/lib/R/library ## ## ────────────────────────────────────────────────────────────────────────────── "],["references.html", "Chapter 4 References", " Chapter 4 References "],["404.html", "Page not found", " Page not found The page you requested cannot be found (perhaps it was moved or renamed). You may want to try searching to find the page's new location, or use the table of contents to find the page you are looking for. "]]
+[["index.html", "Introduction to Python About this Course 0.1 Curriculum 0.2 Target Audience 0.3 Learning Objectives 0.4 Offerings", " Introduction to Python August, 2024 About this Course 0.1 Curriculum The course covers fundamentals of Python, a high-level programming language, and use it to wrangle data for analysis and visualization. 0.2 Target Audience The course is intended for researchers who want to learn coding for the first time with a data science application via the Python language. This course is also appropriate for folks who have explored data science or programming on their own and want to focus on some fundamentals. 0.3 Learning Objectives Analyze Tidy datasets in the Python programming language via data subsetting, joining, and transformations. Evaluate summary statistics and data visualization to understand scientific questions. Describe how the Python programming environment interpret complex expressions made out of functions, operations, and data structures, in a step-by-step way. Apply problem solving strategies to debug broken code. 0.4 Offerings This course is taught on a regular basis at Fred Hutch Cancer Center through the Data Science Lab. Announcements of course offering can be found here. "],["intro-to-computing.html", "Chapter 1 Intro to Computing 1.1 Goals of the course 1.2 What is a computer program? 1.3 A programming language has following elements: 1.4 Google Colab Setup 1.5 Grammar Structure 1: Evaluation of Expressions 1.6 Grammar Structure 2: Storing data types in the Variable Environment 1.7 Grammar Structure 3: Evaluation of Functions 1.8 Tips on writing your first code 1.9 Exercises", " Chapter 1 Intro to Computing Welcome to Introduction to Python! Each week, we cover a chapter, which consists of a lesson and exercise. In our first week together, we will look at big conceptual themes in programming, see how code is run, and learn some basic grammar structures of programming. 1.1 Goals of the course In the next 6 weeks, we will explore: Fundamental concepts in high-level programming languages (Python, R, Julia, etc.) that is transferable: How do programs run, and how do we solve problems using functions and data structures? Beginning of data science fundamentals: How do you translate your scientific question to a data wrangling problem and answer it? Data science workflow. Image source: R for Data Science. Find a nice balance between the two throughout the course: we will try to reproduce a figure from a scientific publication using new data. 1.2 What is a computer program? A sequence of instructions to manipulate data for the computer to execute. A series of translations: English <-> Programming Code for Interpreter <-> Machine Code for Central Processing Unit (CPU) We will focus on English <-> Programming Code for Python Interpreter in this class. More importantly: How we organize ideas <-> Instructing a computer to do something. 1.3 A programming language has following elements: Grammar structure to construct expressions; combining expressions to create more complex expressions Encapsulate complex expressions via functions to create modular and reusable tasks Encapsulate complex data via data structures to allow efficient manipulation of data 1.4 Google Colab Setup Google Colab is a Integrated Development Environment (IDE) on a web browser. Think about it as Microsoft Word to a plain text editor. It provides extra bells and whistles to using Python that is easier for the user. Let’s open up the KRAS analysis in Google Colab. If you are taking this course while it is in session, the project name is probably named “KRAS Demo” in your Google Classroom workspace. If you are taking this course on your own time, you can view it here. Today, we will pay close attention to: Python Console (“Executions”): Open it via View -> Executed code history. You give it one line of Python code, and the console executes that single line of code; you give it a single piece of instruction, and it executes it for you. Notebook: in the central panel of the website, you will see Python code interspersed with word document text. This is called a Python Notebook (other similar services include Jupyter Notebook, iPython Notebook), which has chunks of plain text and Python code, and it helps us understand better the code we are writing. Variable Environment: Open it by clicking on the “{x}” button on the left-hand panel. Often, your code will store information in the Variable Environment, so that information can be reused. For instance, we often load in data and store it in the Variable Environment, and use it throughout rest of your Python code. The first thing we will do is see the different ways we can run Python code. You can do the following: Type something into the Python Console (Execution) and click the arrow button, such as 2+2. The Python Console will run it and give you an output. Look through the Python Notebook, and when you see a chunk of Python Code, click the arrow button. It will copy the Python code chunk to the Python Console and run all of it. You will likely see variables created in the Variables panel as you load in and manipulate data. Run every single Python code chunk via Runtime -> Run all. Remember that the order that you run your code matters in programming. Your final product would be the result of Option 3, in which you run every Python code chunk from start to finish. However, sometimes it is nice to try out smaller parts of your code via Options 1 or 2. But you will be at risk of running your code out of order! To create your own content in the notebook, click on a section you want to insert content, and then click on “+ Code” or “+ Text” to add Python code or text, respectively. Python Notebook is great for data science work, because: It encourages reproducible data analysis, when you run your analysis from start to finish. It encourages excellent documentation, as you can have code, output from code, and prose combined together. It is flexible to use other programming languages, such as R. Now, we will get to the basics of programming grammar. 1.5 Grammar Structure 1: Evaluation of Expressions Expressions are be built out of operations or functions. Functions and operations take in data types, do something with them, and return another data type. We can combine multiple expressions together to form more complex expressions: an expression can have other expressions nested inside it. For instance, consider the following expressions entered to the Python Console: 18 + 21 ## 39 max(18, 21) ## 21 max(18 + 21, 65) ## 65 18 + (21 + 65) ## 104 len("ATCG") ## 4 Here, our input data types to the operation are integer in lines 1-4 and our input data type to the function is string in line 5. We will go over common data types shortly. Operations are just functions in hiding. We could have written: from operator import add add(18, 21) ## 39 add(18, add(21, 65)) ## 104 Remember that the Python language is supposed to help us understand what we are writing in code easily, lending to readable code. Therefore, it is sometimes useful to come up with operations that is easier to read. (Most functions in Python are stored in a collection of functions called modules that needs to be loaded. The import statement gives us permission to access the functions in the module “operator”.) 1.5.1 Data types Here are some common data types we will be using in this course. Data type name Data type shorthand Examples Integer int 2, 4 Float float 3.5, -34.1009 String str “hello”, “234-234-8594” Boolean bool True, False 1.5.2 Function machine schema A nice way to summarize this first grammar structure is using the function machine schema, way back from algebra class: Function machine from algebra class. Here are some aspects of this schema to pay attention to: A programmer should not need to know how the function or operation is implemented in order to use it - this emphasizes abstraction and modular thinking, a foundation in any programming language. A function can have different kinds of inputs and outputs - it doesn’t need to be numbers. In the len() function, the input is a String, and the output is an Integer. We will see increasingly complex functions with all sorts of different inputs and outputs. 1.6 Grammar Structure 2: Storing data types in the Variable Environment To build up a computer program, we need to store our returned data type from our expression somewhere for downstream use. We can assign a variable to it as follows: x = 18 + 21 If you enter this in the Console, you will see that in the Variable Environment, the variable x has a value of 39. 1.6.1 Execution rule for variable assignment Evaluate the expression to the right of =. Bind variable to the left of = to the resulting value. The variable is stored in the Variable Environment. The Variable Environment is where all the variables are stored, and can be used for an expression anytime once it is defined. Only one unique variable name can be defined. The variable is stored in the working memory of your computer, Random Access Memory (RAM). This is temporary memory storage on the computer that can be accessed quickly. Typically a personal computer has 8, 16, 32 Gigabytes of RAM. Look, now x can be reused downstream: x - 2 ## 37 y = x * 2 It is quite common for programmers to have to look up the data type of a variable while they are coding. To learn about the data type of a variable, use the type() function on any variable in Python: type(y) ## <class 'int'> We should give useful variable names so that we know what to expect! If you are working with numerical sales data, consider num_sales instead of y. 1.7 Grammar Structure 3: Evaluation of Functions Let’s look at functions a little bit more formally: A function has a function name, arguments, and returns a data type. 1.7.1 Execution rule for functions: Evaluate the function by its arguments if there’s any, and if the arguments are functions or contains operations, evaluate those functions or operations first. The output of functions is called the returned value. Often, we will use multiple functions in a nested way, and it is important to understand how the Python console understand the order of operation. We can also use paranthesis to change the order of operation. Think about what the Python is going to do step-by–step in the lines of code below: max(len("hello"), 4) ## 5 (len("pumpkin") - 8) * 2 ## -2 If we don’t know how to use a function, such as pow(), we can ask for help: ?pow pow(base, exp, mod=None) Equivalent to base**exp with 2 arguments or base**exp % mod with 3 arguments Some types, such as ints, are able to use a more efficient algorithm when invoked using the three argument form. This shows the function takes in three input arguments: base, exp, and mod=None. When an argument has an assigned value of mod=None, that means the input argument already has a value, and you don’t need to specify anything, unless you want to. The following ways are equivalent ways of using the pow() function: pow(2, 3) ## 8 pow(base=2, exp=3) ## 8 pow(exp=3, base=2) ## 8 but this will give you something different: pow(3, 2) ## 9 And there is an operational equivalent: 2 ** 3 ## 8 We will mostly look at functions with input arguments and return types in this course, but not all functions need to have input arguments and output return. Here are some varieties of functions to stretch your horizons. Function call What it takes in What it does Returns pow(a, b) integer a, integer b Raises a to the bth power. Integer print(x) any data type x Prints out the value of x to the console. None datetime.now() Nothing Gets the current time. String 1.8 Tips on writing your first code Computer = powerful + stupid Computers are excellent at doing something specific over and over again, but is extremely rigid and lack flexibility. Here are some tips that is helpful for beginners: Write incrementally, test often Check your assumptions, especially using new functions, operations, and new data types. Live environments are great for testing, but not great for reproducibility. Ask for help! To get more familiar with the errors Python gives you, take a look at this summary of Python error messages. 1.9 Exercises Exercise for week 1 can be found here. "],["working-with-data-structures.html", "Chapter 2 Working with data structures 2.1 Lists 2.2 Objects in Python 2.3 Dataframes 2.4 Exercises", " Chapter 2 Working with data structures In our second lesson, we start to look at two data structures, Lists and Dataframes, that can handle a large amount of data for analysis. 2.1 Lists In the first exercise, you started to explore data structures, which store information about data types. You explored lists, which is an ordered collection of data types or data structures. Each element of a list contains a data type or another data structure. We can now store a vast amount of information in a list, and assign it to a single variable. Even more, we can use operations and functions on a list, modifying many elements within the list at once! This makes analyzing data much more scalable and less repetitive. We create a list via the bracket [ ] operation. staff = ["chris", "ted", "jeff"] chrNum = [2, 3, 1, 2, 2] mixedList = [False, False, False, "A", "B", 92] 2.1.1 Subsetting lists To access an element of a list, you can use the bracket notation [ ] to access the elements of the list. We simply access an element via the “index” number - the location of the data within the list. Here’s the tricky thing about the index number: it starts at 0! 1st element of chrNum: chrNum[0] 2nd element of chrNum: chrNum[1] … 5th element of chrNum: chrNum[4] With subsetting, you can modify elements of a list or use the element of a list as part of an expression. 2.1.2 Subsetting multiple elements of lists Suppose you want to access multiple elements of a list, such as accessing the first three elements of chrNum. You would use the slice operator :, which specifies: the index number to start the index number to stop, plus one. If you want to access the first three elements of chrNum: chrNum[0:3] ## [2, 3, 1] The first element’s index number is 0, the third element’s index number is 2, plus 1, which is 3. If you want to access the second and third elements of chrNum: chrNum[1:3] ## [3, 1] If you want to access everything but the first three elements of chrNum: chrNum[3:] ## [2, 2] Here, the stop index number was not specificed. When the start or stop index is not specified, it implies that you are subsetting starting the from the beginning of the list or subsetting to the end of the list, respectively: chrNum[:3] ## [2, 3, 1] chrNum[3:] ## [2, 2] There are other popular uses of the slice operator :, such as negative indicies to count from the end of a list, or subsetting with a fixed increment. You can find more discussion of list slicing here. 2.2 Objects in Python The list data structure has an organization and functionality that metaphorically represents a pen-and-paper list in our physical world. Like a physical object, we have examined: What does it contain (in terms of data)? What can it do (in terms of operations and functions)? And if it “makes sense” to us, then it is well-designed. The list data structure we have been working with is an example of an Object. The definition of an object allows us to ask the questions above: what does it contain, and what can it do? It is an organizational tool for a collection of data and functions that we can relate to, like a physical object. Formally, an object contains the following: Value that holds the essential data for the object. Attributes that store additional data for the object. Functions called Methods that can be used on the object. This organizing structure on an object applies to pretty much all Python data types and data structures. Let’s see how this applies to the list: Value: the contents of the list, such as [2, 3, 4]. Attributes that store additional values: Not relevant for lists. Methods that can be used on the object: chrNum.count(2) counts the number of instances 2 appears as an element of chrNum. Object methods are functions that does something with the object you are using it on. You should think about chrNum.count(2) as a function that takes in chrNum and 2 as inputs. If you want to use the count function on list mixedList, you would use mixedList.count(x). Here are some more examples of methods with lists: Function method What it takes in What it does Returns chrNum.count(x) list chrNum, data type x Counts the number of instances x appears as an element of chrNum. Integer chrNum.append(x) list chrNum, data type x Appends x to the end of the chrNum. None (but chrNum is modified!) chrNum.sort() list chrNum Sorts chrNum by ascending order. None (but chrNum is modified!) chrNum.reverse() list chrNum Reverses the order of chrNum. None (but chrNum is modified!) 2.3 Dataframes A Dataframe is a two-dimensional data structure that stores data like a spreadsheet does. The Dataframe data structure is found within a Python module called “Pandas”. A Python module is an organized collection of functions and data structures. The import statement below gives us permission to access the “Pandas” module via the variable pd. To load in a Dataframe from existing spreadsheet data, we use the function pd.read_csv(): import pandas as pd metadata = pd.read_csv("classroom_data/metadata.csv") type(metadata) ## <class 'pandas.core.frame.DataFrame'> There is a similar function pd.read_excel() for loading in Excel spreadsheets. Let’s investigate the Dataframe as an object: What does a Dataframe contain (in terms of data)? What can a Dataframe do (in terms of operations and functions)? 2.3.1 What does a Dataframe contain (in terms of data)? We first take a look at the contents: metadata ## ModelID ... OncotreeLineage ## 0 ACH-000001 ... Ovary/Fallopian Tube ## 1 ACH-000002 ... Myeloid ## 2 ACH-000003 ... Bowel ## 3 ACH-000004 ... Myeloid ## 4 ACH-000005 ... Myeloid ## ... ... ... ... ## 1859 ACH-002968 ... Esophagus/Stomach ## 1860 ACH-002972 ... Esophagus/Stomach ## 1861 ACH-002979 ... Esophagus/Stomach ## 1862 ACH-002981 ... Esophagus/Stomach ## 1863 ACH-003071 ... Lung ## ## [1864 rows x 30 columns] It looks like there are 1864 rows and 30 columns in this Dataframe, and when we display it it shows some of the data. We can look at specific columns by looking at attributes via the dot operation. We can also look at the columns via the bracket operation. metadata.ModelID ## 0 ACH-000001 ## 1 ACH-000002 ## 2 ACH-000003 ## 3 ACH-000004 ## 4 ACH-000005 ## ... ## 1859 ACH-002968 ## 1860 ACH-002972 ## 1861 ACH-002979 ## 1862 ACH-002981 ## 1863 ACH-003071 ## Name: ModelID, Length: 1864, dtype: object metadata['ModelID'] ## 0 ACH-000001 ## 1 ACH-000002 ## 2 ACH-000003 ## 3 ACH-000004 ## 4 ACH-000005 ## ... ## 1859 ACH-002968 ## 1860 ACH-002972 ## 1861 ACH-002979 ## 1862 ACH-002981 ## 1863 ACH-003071 ## Name: ModelID, Length: 1864, dtype: object The names of all columns is stored as an attribute, which can be accessed via the dot operation. metadata.columns ## Index(['ModelID', 'PatientID', 'CellLineName', 'StrippedCellLineName', 'Age', ## 'SourceType', 'SangerModelID', 'RRID', 'DepmapModelType', 'AgeCategory', ## 'GrowthPattern', 'LegacyMolecularSubtype', 'PrimaryOrMetastasis', ## 'SampleCollectionSite', 'Sex', 'SourceDetail', 'LegacySubSubtype', ## 'CatalogNumber', 'CCLEName', 'COSMICID', 'PublicComments', ## 'WTSIMasterCellID', 'EngineeredModel', 'TreatmentStatus', ## 'OnboardedMedia', 'PlateCoating', 'OncotreeCode', 'OncotreeSubtype', ## 'OncotreePrimaryDisease', 'OncotreeLineage'], ## dtype='object') The number of rows and columns are also stored as an attribute: metadata.shape ## (1864, 30) 2.3.2 What can a Dataframe do (in terms of operations and functions)? We can use the head() and tail() functions to look at the first few rows and last few rows of metadata, respectively: metadata.head() ## ModelID PatientID ... OncotreePrimaryDisease OncotreeLineage ## 0 ACH-000001 PT-gj46wT ... Ovarian Epithelial Tumor Ovary/Fallopian Tube ## 1 ACH-000002 PT-5qa3uk ... Acute Myeloid Leukemia Myeloid ## 2 ACH-000003 PT-puKIyc ... Colorectal Adenocarcinoma Bowel ## 3 ACH-000004 PT-q4K2cp ... Acute Myeloid Leukemia Myeloid ## 4 ACH-000005 PT-q4K2cp ... Acute Myeloid Leukemia Myeloid ## ## [5 rows x 30 columns] metadata.tail() ## ModelID PatientID ... OncotreePrimaryDisease OncotreeLineage ## 1859 ACH-002968 PT-pjhrsc ... Esophagogastric Adenocarcinoma Esophagus/Stomach ## 1860 ACH-002972 PT-dkXZB1 ... Esophagogastric Adenocarcinoma Esophagus/Stomach ## 1861 ACH-002979 PT-lyHTzo ... Esophagogastric Adenocarcinoma Esophagus/Stomach ## 1862 ACH-002981 PT-Z9akXf ... Esophagogastric Adenocarcinoma Esophagus/Stomach ## 1863 ACH-003071 PT-LAGmLq ... Lung Neuroendocrine Tumor Lung ## ## [5 rows x 30 columns] Both of these functions (without input arguments) are considered as methods: they are functions that does something with the Dataframe you are using it on. You should think about metadata.head() as a function that takes in metadata as an input. If we had another Dataframe called my_data and you want to use the same function, you will have to say my_data.head(). 2.3.2.1 Subsetting Dataframes Perhaps the most important operation you will can do with Dataframes is subsetting them. There are two ways to do it. The first way is to subset by numerical indicies, exactly like how we did for lists. You will use the iloc and bracket operations, and you give two slices: one for the row, and one for the column. Let’s start with a small dataframe to see how it works before returning to metadata: df = pd.DataFrame(data={'status': ["treated", "untreated", "untreated", "discharged", "treated"], 'age_case': [25, 43, 21, 65, 7], 'age_control': [49, 20, 32, 25, 32]}) df ## status age_case age_control ## 0 treated 25 49 ## 1 untreated 43 20 ## 2 untreated 21 32 ## 3 discharged 65 25 ## 4 treated 7 32 Here is how the dataframe looks like with the row and column index numbers: Subset the second to fourth rows, and the first two columns: Now, back to metadata dataframe: Subset the first 5 rows, and first two columns: metadata.iloc[:5, :2] ## ModelID PatientID ## 0 ACH-000001 PT-gj46wT ## 1 ACH-000002 PT-5qa3uk ## 2 ACH-000003 PT-puKIyc ## 3 ACH-000004 PT-q4K2cp ## 4 ACH-000005 PT-q4K2cp If we want a custom slice that is not sequential, we can use an integer list. Subset the last 5 rows, and the 1st and 10 and 21th column: metadata.iloc[5:, [1, 10, 21]] ## PatientID GrowthPattern WTSIMasterCellID ## 5 PT-ej13Dz Suspension 2167.0 ## 6 PT-NOXwpH Adherent 569.0 ## 7 PT-fp8PeY Adherent 1806.0 ## 8 PT-puKIyc Adherent 2104.0 ## 9 PT-AR7W9o Adherent NaN ## ... ... ... ... ## 1859 PT-pjhrsc Organoid NaN ## 1860 PT-dkXZB1 Organoid NaN ## 1861 PT-lyHTzo Organoid NaN ## 1862 PT-Z9akXf Organoid NaN ## 1863 PT-LAGmLq Suspension NaN ## ## [1859 rows x 3 columns] When we subset via numerical indicies, it’s called explicit subsetting. This is a great way to start thinking about subsetting your dataframes for analysis, but explicit subsetting can lead to some inconsistencies in the long run. For instance, suppose your collaborator added a new cell line to the metadata and changed the order of the column. Then your code to subset the last 5 rows and the columns will get you a different answer once the spreadsheet is changed. The second way is to subset by the column name and comparison operators, also known as implicit subsetting. This is much more robust in data analysis practice. You will learn about it next week! 2.4 Exercises Exercise for week 2 can be found here. "],["data-wrangling-part-1.html", "Chapter 3 Data Wrangling, Part 1 3.1 Tidy Data 3.2 Our working Tidy Data: DepMap Project 3.3 Transform: “What do you want to do with this Dataframe”? 3.4 Summary Statistics 3.5 Simple data visualization 3.6 Exercises", " Chapter 3 Data Wrangling, Part 1 From our first two lessons, we are now equipped with enough fundamental programming skills to apply it to various steps in the data science workflow, which is a natural cycle that occurs in data analysis. Data science workflow. Image source: R for Data Science. For the rest of the course, we focus on Transform and Visualize with the assumption that our data is in a nice, “Tidy format”. First, we need to understand what it means for a data to be “Tidy”. 3.1 Tidy Data Here, we describe a standard of organizing data. It is important to have standards, as it facilitates a consistent way of thinking about data organization and building tools (functions) that make use of that standard. The principles of tidy data, developed by Hadley Wickham: Each variable must have its own column. Each observation must have its own row. Each value must have its own cell. If you want to be technical about what variables and observations are, Hadley Wickham describes: A variable contains all values that measure the same underlying attribute (like height, temperature, duration) across units. An observation contains all values measured on the same unit (like a person, or a day, or a race) across attributes. A tidy dataframe. Image source: R for Data Science. 3.2 Our working Tidy Data: DepMap Project The Dependency Map project is a multi-omics profiling of cancer cell lines combined with functional assays such as CRISPR and drug sensitivity to help identify cancer vulnerabilities and drug targets. Here are some of the data that we have public access to. We have been looking at the metadata since last session. Metadata Somatic mutations Gene expression Drug sensitivity CRISPR knockout and more… Let’s load these datasets in, and see how these datasets fit the definition of Tidy data: import pandas as pd metadata = pd.read_csv("classroom_data/metadata.csv") mutation = pd.read_csv("classroom_data/mutation.csv") expression = pd.read_csv("classroom_data/expression.csv") metadata.head() ## ModelID PatientID ... OncotreePrimaryDisease OncotreeLineage ## 0 ACH-000001 PT-gj46wT ... Ovarian Epithelial Tumor Ovary/Fallopian Tube ## 1 ACH-000002 PT-5qa3uk ... Acute Myeloid Leukemia Myeloid ## 2 ACH-000003 PT-puKIyc ... Colorectal Adenocarcinoma Bowel ## 3 ACH-000004 PT-q4K2cp ... Acute Myeloid Leukemia Myeloid ## 4 ACH-000005 PT-q4K2cp ... Acute Myeloid Leukemia Myeloid ## ## [5 rows x 30 columns] mutation.head() ## ModelID CACNA1D_Mut CYP2D6_Mut ... CCDC28A_Mut C1orf194_Mut U2AF1_Mut ## 0 ACH-000001 False False ... False False False ## 1 ACH-000002 False False ... False False False ## 2 ACH-000004 False False ... False False False ## 3 ACH-000005 False False ... False False False ## 4 ACH-000006 False False ... False False False ## ## [5 rows x 540 columns] expression.head() ## ModelID ENPP4_Exp CREBBP_Exp ... OR5D13_Exp C2orf81_Exp OR8S1_Exp ## 0 ACH-001113 2.280956 4.094236 ... 0.0 1.726831 0.0 ## 1 ACH-001289 3.622930 3.606442 ... 0.0 0.790772 0.0 ## 2 ACH-001339 0.790772 2.970854 ... 0.0 0.575312 0.0 ## 3 ACH-001538 3.485427 2.801159 ... 0.0 1.077243 0.0 ## 4 ACH-000242 0.879706 3.327687 ... 0.0 0.722466 0.0 ## ## [5 rows x 536 columns] Dataframe The observation is Some variables are Some values are metadata Cell line ModelID, Age, OncotreeLineage “ACH-000001”, 60, “Myeloid” expression Cell line KRAS_Exp 2.4, .3 mutation Cell line KRAS_Mut TRUE, FALSE 3.3 Transform: “What do you want to do with this Dataframe”? Remember that a major theme of the course is about: How we organize ideas <-> Instructing a computer to do something. Until now, we haven’t focused too much on how we organize our scientific ideas to interact with what we can do with code. Let’s pivot to write our code driven by our scientific curiosity. After we are sure that we are working with Tidy data, we can ponder how we want to transform our data that satisfies our scientific question. We will look at several ways we can transform Tidy data, starting with subsetting columns and rows. Here’s a starting prompt: In the metadata dataframe, which rows would you subset for and columns would you subset for that relate to a scientific question? We have been using explicit subsetting with numerical indicies, such as “I want to filter for rows 20-50 and select columns 2 and 8”. We are now going to switch to implicit subsetting in which we describe the subsetting criteria via comparision operators and column names, such as: “I want to subset for rows such that the OncotreeLineage is breast cancer and subset for columns Age and Sex.” Notice that when we subset for rows in an implicit way, we formulate our criteria in terms of the columns.This is because we are guaranteed to have column names in Dataframes, but not row names. 3.3.0.1 Let’s convert our implicit subsetting criteria into code! To subset for rows implicitly, we will use the conditional operators on Dataframe columns you used in Exercise 2. To formulate a conditional operator expression that OncotreeLineage is breast cancer: metadata['OncotreeLineage'] == "Lung" ## 0 False ## 1 False ## 2 False ## 3 False ## 4 False ## ... ## 1859 False ## 1860 False ## 1861 False ## 1862 False ## 1863 True ## Name: OncotreeLineage, Length: 1864, dtype: bool Then, we will use the .loc operation (which is different than .iloc operation!) and subsetting brackets to subset rows and columns Age and Sex at the same time: metadata.loc[metadata['OncotreeLineage'] == "Lung", ["Age", "Sex"]] ## Age Sex ## 10 39.0 Female ## 13 44.0 Male ## 19 55.0 Female ## 27 39.0 Female ## 28 45.0 Male ## ... ... ... ## 1745 52.0 Male ## 1819 84.0 Male ## 1820 57.0 Female ## 1822 53.0 Male ## 1863 62.0 Male ## ## [241 rows x 2 columns] What’s going on here? The first component of the subset, metadata['OncotreeLineage'] == \"Lung\", subsets for the rows. It gives us a column of True and False values, and we keep rows that correspond to True values. Then, we specify the column names we want to subset for via a list. Here’s another example: df = pd.DataFrame(data={'status': ["treated", "untreated", "untreated", "discharged", "treated"], 'age_case': [25, 43, 21, 65, 7], 'age_control': [49, 20, 32, 25, 32]}) df ## status age_case age_control ## 0 treated 25 49 ## 1 untreated 43 20 ## 2 untreated 21 32 ## 3 discharged 65 25 ## 4 treated 7 32 “I want to subset for rows such that the status is”treated” and subset for columns status and age_case.” df.loc[df.status == "treated", ["status", "age_case"]] ## status age_case ## 0 treated 25 ## 4 treated 7 3.4 Summary Statistics Now that your Dataframe has be transformed based on your scientific question, you can start doing some analysis on it! A common data science task is to examine summary statistics of a dataset, which summarizes the all the values from a variable in a numeric summary, such as mean, median, or mode. If we look at the data structure of a Dataframe’s column, it is actually not a List, but an object called Series. It has methods can compute summary statistics for us. Let’s take a look at a few popular examples: Function method What it takes in What it does Returns metadata.Age.mean() metadata.Age as a numeric Series Computes the mean value of the Age column. Float (NumPy) metadata['Age'].median() metadata['Age'] as a numeric Series Computes the median value of the Age column. Float (NumPy) metadata.Age.max() metadata.Age as a numeric Series Computes the max value of the Age column. Float (NumPy) metadata.OncotreeSubtype.value_counts() metadata.OncotreeSubtype as a string Series Creates a frequency table of all unique elements in OncotreeSubtype column. Series Let’s try it out, with some nice print formatting: print("Mean value of Age column:", metadata['Age'].mean()) ## Mean value of Age column: 47.45187165775401 print("Frequency of column", metadata.OncotreeLineage.value_counts()) ## Frequency of column OncotreeLineage ## Lung 241 ## Lymphoid 209 ## CNS/Brain 123 ## Skin 118 ## Esophagus/Stomach 95 ## Breast 92 ## Bowel 87 ## Head and Neck 81 ## Myeloid 77 ## Bone 75 ## Ovary/Fallopian Tube 74 ## Pancreas 65 ## Kidney 64 ## Peripheral Nervous System 55 ## Soft Tissue 54 ## Uterus 41 ## Fibroblast 41 ## Biliary Tract 40 ## Bladder/Urinary Tract 39 ## Normal 39 ## Pleura 35 ## Liver 28 ## Cervix 25 ## Eye 19 ## Thyroid 18 ## Prostate 14 ## Vulva/Vagina 5 ## Ampulla of Vater 4 ## Testis 4 ## Adrenal Gland 1 ## Other 1 ## Name: count, dtype: int64 Notice that the output of some of these methods are Float (NumPy). This refers to a Python Object called NumPy that is extremely popular for scientific computing, but we’re not focused on that in this course.) 3.5 Simple data visualization We will dedicate extensive time later this course to talk about data visualization, but the Dataframe’s column, Series, has a method called .plot() that can help us make simple plots for one variable. The .plot() method will by default make a line plot, but it is not necessary the plot style we want, so we can give the optional argument kind a String value to specify the plot style. We use it for making a histogram or bar plot. Plot style Useful for kind = Code Histogram Numerics “hist” metadata.Age.plot(kind = \"hist\") Bar plot Strings “bar” metadata.OncotreeSubtype.value_counts().plot(kind = \"bar\") Let’s look at a histogram: import matplotlib.pyplot as plt plt.figure() metadata.Age.plot(kind = "hist") plt.show() Let’s look at a bar plot: plt.figure() metadata.OncotreeLineage.value_counts().plot(kind = "bar") plt.show() (The plt.figure() and plt.show() functions are used to render the plots on the website, but you don’t need to use it for your exercises - yet. We will discuss this in more detail during our week of data visualization.) 3.5.0.1 Chained function calls Let’s look at our bar plot syntax more carefully. We start with the column metadata.OncotreeLineage, and then we first use the method .value_counts() to get a Series of a frequency table. Then, we take the frequency table Series and use the .plot() method. It is quite common in Python to have multiple “chained” function calls, in which the output of .value_counts() is used for the input of .plot() all in one line of code. It takes a bit of time to get used to this! Here’s another example of a chained function call, which looks quite complex, but let’s break it down: plt.figure() metadata.loc[metadata.AgeCategory == "Adult", ].OncotreeLineage.value_counts().plot(kind="bar") plt.show() We first take the entire metadata and do some subsetting, which outputs a Dataframe. We access the OncotreeLineage column, which outputs a Series. We use the method .value_counts(), which outputs a Series. We make a plot out of it! We could have, alternatively, done this in several lines of code: plt.figure() metadata_subset = metadata.loc[metadata.AgeCategory == "Adult", ] metadata_subset_lineage = metadata_subset.OncotreeLineage lineage_freq = metadata_subset_lineage.value_counts() lineage_freq.plot(kind = "bar") plt.show() These are two different styles of code, but they do the exact same thing. It’s up to you to decide what is easier for you to understand. 3.6 Exercises Exercise for week 3 can be found here. "],["about-the-authors.html", "About the Authors", " About the Authors These credits are based on our course contributors table guidelines. Credits Names Pedagogy Lead Content Instructor(s) FirstName LastName Lecturer(s) (include chapter name/link in parentheses if only for specific chapters) - make new line if more than one chapter involved Delivered the course in some way - video or audio Content Author(s) (include chapter name/link in parentheses if only for specific chapters) - make new line if more than one chapter involved If any other authors besides lead instructor Content Contributor(s) (include section name/link in parentheses) - make new line if more than one section involved Wrote less than a chapter Content Editor(s)/Reviewer(s) Checked your content Content Director(s) Helped guide the content direction Content Consultants (include chapter name/link in parentheses or word “General”) - make new line if more than one chapter involved Gave high level advice on content Acknowledgments Gave small assistance to content but not to the level of consulting Production Content Publisher(s) Helped with publishing platform Content Publishing Reviewer(s) Reviewed overall content and aesthetics on publishing platform Technical Course Publishing Engineer(s) Helped with the code for the technical aspects related to the specific course generation Template Publishing Engineers Candace Savonen, Carrie Wright, Ava Hoffman Publishing Maintenance Engineer Candace Savonen Technical Publishing Stylists Carrie Wright, Ava Hoffman, Candace Savonen Package Developers (ottrpal) Candace Savonen, John Muschelli, Carrie Wright Art and Design Illustrator(s) Created graphics for the course Figure Artist(s) Created figures/plots for course Videographer(s) Filmed videos Videography Editor(s) Edited film Audiographer(s) Recorded audio Audiography Editor(s) Edited audio recordings Funding Funder(s) Institution/individual who funded course including grant number Funding Staff Staff members who help with funding ## ─ Session info ─────────────────────────────────────────────────────────────── ## setting value ## version R version 4.3.2 (2023-10-31) ## os Ubuntu 22.04.4 LTS ## system x86_64, linux-gnu ## ui X11 ## language (EN) ## collate en_US.UTF-8 ## ctype en_US.UTF-8 ## tz Etc/UTC ## date 2024-08-22 ## pandoc 3.1.1 @ /usr/local/bin/ (via rmarkdown) ## ## ─ Packages ─────────────────────────────────────────────────────────────────── ## package * version date (UTC) lib source ## askpass 1.2.0 2023-09-03 [1] RSPM (R 4.3.0) ## bookdown 0.39.1 2024-06-11 [1] Github (rstudio/bookdown@f244cf1) ## bslib 0.6.1 2023-11-28 [1] RSPM (R 4.3.0) ## cachem 1.0.8 2023-05-01 [1] RSPM (R 4.3.0) ## cli 3.6.2 2023-12-11 [1] RSPM (R 4.3.0) ## devtools 2.4.5 2022-10-11 [1] RSPM (R 4.3.0) ## digest 0.6.34 2024-01-11 [1] RSPM (R 4.3.0) ## ellipsis 0.3.2 2021-04-29 [1] RSPM (R 4.3.0) ## evaluate 0.23 2023-11-01 [1] RSPM (R 4.3.0) ## fansi 1.0.6 2023-12-08 [1] RSPM (R 4.3.0) ## fastmap 1.1.1 2023-02-24 [1] RSPM (R 4.3.0) ## fs 1.6.3 2023-07-20 [1] RSPM (R 4.3.0) ## glue 1.7.0 2024-01-09 [1] RSPM (R 4.3.0) ## hms 1.1.3 2023-03-21 [1] RSPM (R 4.3.0) ## htmltools 0.5.7 2023-11-03 [1] RSPM (R 4.3.0) ## htmlwidgets 1.6.4 2023-12-06 [1] RSPM (R 4.3.0) ## httpuv 1.6.14 2024-01-26 [1] RSPM (R 4.3.0) ## httr 1.4.7 2023-08-15 [1] RSPM (R 4.3.0) ## jquerylib 0.1.4 2021-04-26 [1] RSPM (R 4.3.0) ## jsonlite 1.8.8 2023-12-04 [1] RSPM (R 4.3.0) ## knitr 1.47.3 2024-06-11 [1] Github (yihui/knitr@e1edd34) ## later 1.3.2 2023-12-06 [1] RSPM (R 4.3.0) ## lifecycle 1.0.4 2023-11-07 [1] RSPM (R 4.3.0) ## magrittr 2.0.3 2022-03-30 [1] RSPM (R 4.3.0) ## memoise 2.0.1 2021-11-26 [1] RSPM (R 4.3.0) ## mime 0.12 2021-09-28 [1] RSPM (R 4.3.0) ## miniUI 0.1.1.1 2018-05-18 [1] RSPM (R 4.3.0) ## openssl 2.1.1 2023-09-25 [1] RSPM (R 4.3.0) ## ottrpal 1.2.1 2024-06-11 [1] Github (jhudsl/ottrpal@828539f) ## pillar 1.9.0 2023-03-22 [1] RSPM (R 4.3.0) ## pkgbuild 1.4.3 2023-12-10 [1] RSPM (R 4.3.0) ## pkgconfig 2.0.3 2019-09-22 [1] RSPM (R 4.3.0) ## pkgload 1.3.4 2024-01-16 [1] RSPM (R 4.3.0) ## profvis 0.3.8 2023-05-02 [1] RSPM (R 4.3.0) ## promises 1.2.1 2023-08-10 [1] RSPM (R 4.3.0) ## purrr 1.0.2 2023-08-10 [1] RSPM (R 4.3.0) ## R6 2.5.1 2021-08-19 [1] RSPM (R 4.3.0) ## Rcpp 1.0.12 2024-01-09 [1] RSPM (R 4.3.0) ## readr 2.1.5 2024-01-10 [1] RSPM (R 4.3.0) ## remotes 2.4.2.1 2023-07-18 [1] RSPM (R 4.3.0) ## rlang 1.1.4 2024-06-04 [1] CRAN (R 4.3.2) ## rmarkdown 2.27.1 2024-06-11 [1] Github (rstudio/rmarkdown@e1c93a9) ## sass 0.4.8 2023-12-06 [1] RSPM (R 4.3.0) ## sessioninfo 1.2.2 2021-12-06 [1] RSPM (R 4.3.0) ## shiny 1.8.0 2023-11-17 [1] RSPM (R 4.3.0) ## stringi 1.8.3 2023-12-11 [1] RSPM (R 4.3.0) ## stringr 1.5.1 2023-11-14 [1] RSPM (R 4.3.0) ## tibble 3.2.1 2023-03-20 [1] CRAN (R 4.3.2) ## tzdb 0.4.0 2023-05-12 [1] RSPM (R 4.3.0) ## urlchecker 1.0.1 2021-11-30 [1] RSPM (R 4.3.0) ## usethis 2.2.3 2024-02-19 [1] RSPM (R 4.3.0) ## utf8 1.2.4 2023-10-22 [1] RSPM (R 4.3.0) ## vctrs 0.6.5 2023-12-01 [1] RSPM (R 4.3.0) ## xfun 0.44.4 2024-06-11 [1] Github (yihui/xfun@9da62cc) ## xml2 1.3.6 2023-12-04 [1] RSPM (R 4.3.0) ## xtable 1.8-4 2019-04-21 [1] RSPM (R 4.3.0) ## yaml 2.3.8 2023-12-11 [1] RSPM (R 4.3.0) ## ## [1] /usr/local/lib/R/site-library ## [2] /usr/local/lib/R/library ## ## ────────────────────────────────────────────────────────────────────────────── "],["references.html", "Chapter 4 References", " Chapter 4 References "],["404.html", "Page not found", " Page not found The page you requested cannot be found (perhaps it was moved or renamed). You may want to try searching to find the page's new location, or use the table of contents to find the page you are looking for. "]]
diff --git a/docs/no_toc/working-with-data-structures.html b/docs/no_toc/working-with-data-structures.html
index da3efc3..7d07291 100644
--- a/docs/no_toc/working-with-data-structures.html
+++ b/docs/no_toc/working-with-data-structures.html
@@ -262,15 +262,14 @@ 2.1.2 Subsetting multiple element
## [3, 1]
If you want to access everything but the first three elements of chrNum:
-
+
## [2, 2]
-where len(chrNum) is the length of the list.
-When the start or stop index is not specified, it implies that you are subsetting starting the from the beginning of the list or subsetting to the end of the list, respectively:
+Here, the stop index number was not specificed. When the start or stop index is not specified, it implies that you are subsetting starting the from the beginning of the list or subsetting to the end of the list, respectively:
## [2, 3, 1]
## [2, 2]
-More discussion of list slicing can be found here.
+There are other popular uses of the slice operator :, such as negative indicies to count from the end of a list, or subsetting with a fixed increment. You can find more discussion of list slicing here.
3.4 Summary Statistics
Now that your Dataframe has be transformed based on your scientific question, you can start doing some analysis on it! A common data science task is to examine summary statistics of a dataset, which summarizes the all the values from a variable in a numeric summary, such as mean, median, or mode.
-If we look at the data structre of a Dataframe’s column, it is called a Series. It has methods can compute summary statistics for us. Let’s take a look at a few popular examples:
+If we look at the data structure of a Dataframe’s column, it is actually not a List, but an object called Series. It has methods can compute summary statistics for us. Let’s take a look at a few popular examples:
metadata.Age.mean() |
-metadata.Age as a numeric value |
+metadata.Age as a numeric Series |
Computes the mean value of the Age column. |
Float (NumPy) |
metadata['Age'].median() |
-metadata['Age'] as a numeric value |
+metadata['Age'] as a numeric Series |
Computes the median value of the Age column. |
Float (NumPy) |
metadata.Age.max() |
-metadata.Age as a numeric value |
+metadata.Age as a numeric Series |
Computes the max value of the Age column. |
Float (NumPy) |
metadata.OncotreeSubtype.value_counts() |
-metadata.OncotreeSubtype as a String |
+metadata.OncotreeSubtype as a string Series |
Creates a frequency table of all unique elements in OncotreeSubtype column. |
Series |

