This project is archived for future reference, but no new work is expected in this repository.
This repository can be used as a template for all PACTA workflow repositories. It includes typical workflow scaffolding, and an explanation of good practices for developing a workflow.
When you are starting a new workflow, you may choose to:
- Click "New Repository" on GitHub
- Select this repository as a "template"
After this, you should submit a PR that changes the names in the following files to suit the name of your new repository:
*.RprojfileREADME.md
You can then begin to work on your workflow using the demo.R file as a basis, and document how to run the workflow using the README.md.
These are a handful of guidelines for some best practices in developing a reproducible workflow repository. Please consider this when writing one yourself!
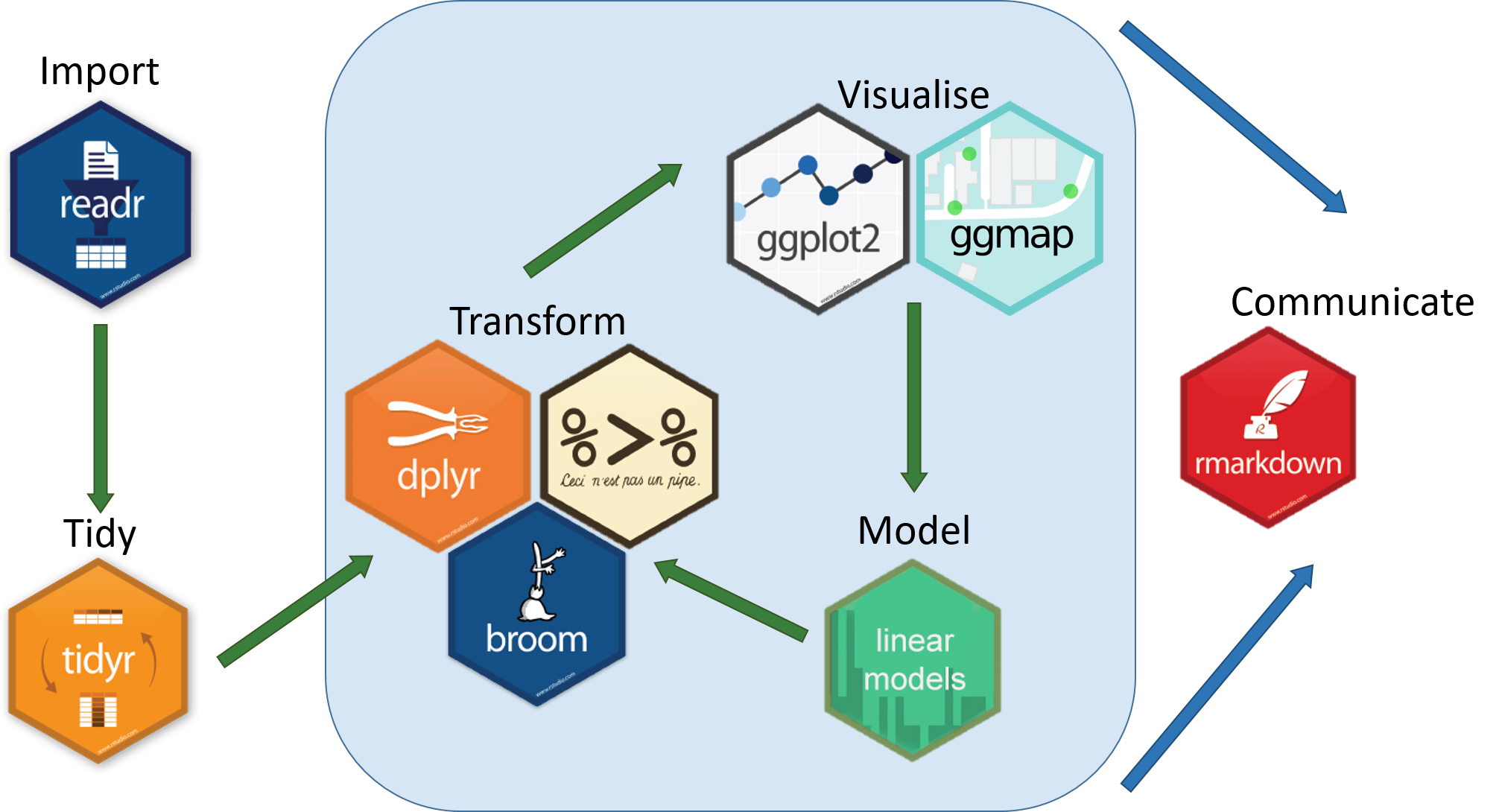
A good way to keep your scripts organized is to think about them in terms of the "data science pipeline". It structures a typical data analysis path as follows:
- Import
- Tidy
- Do the following iteratively:
- Transform
- Visualize
- Model
- Communicate (when you are happy with the output)
In the tidyverse, there are individual packages that can help you with each step of this process:
Don't commit data or other sensitive information to version control. Data should be written to and read from an external source
Create a clear folder structure for your project. Separate code (which can live in this repository) from outputs like: data, figures, and reports.
Consider using R projects (.Rproj) for a more organized environment.
At the beginning of your script, explicitly state all required R packages and their versions. You can use, for example, the renv package to manage package dependencies.
Always try to keep dependencies to a minimum, and remove those that are unused.
If you want to be truly reproducible, then STOP OPENING EXCEL!
Store parameters, configurations, or settings in separate config files (e.g., a config.yml or .env) for easy modification without altering the code.
It is a good idea to document paths here, for example.
Remove unnecessary or temporary files and folders from your script's output to keep things tidy.
If your analysis relies on external data sources, create a separate R script to download and prepare the data automatically.
Develop a checklist to verify that your script meets all reproducibility requirements before sharing it.